Biomolecules interact with each other through the formation and rupture of chemical bonds. Studying these interactions at the single-molecule level contributes significantly to our understanding of biological phenomena. A key challenge associated with such single-molecule studies is the lack of high-throughput methods and convenient study of dynamic processes such as binding and unbinding of molecules.
Now, researchers have used custom-designed DNA leashes that can hold biomolecules such as proteins and exert a pulling force by using acoustic waves to study the strength of the protein-protein bonds. The work involves researchers from the Turing Centre for Living Systems, Marseille Cancer Research Center, and PSL Research University in France. The researchers show proof of concept for the acoustic force method using rapamycin, a clinically used drug that modulates protein-protein interactions in the body.
The groups studied the FKBP12-rapamycin-FRB complex, in which rapamycin modulates the protein-protein interactions. Unlike traditional single-molecule methods in which the interacting molecules are taken apart once a force is exerted, the authors used a junctured-DNA (J-DNA) scaffold that keeps the interacting molecules in close proximity after they are separated, allowing rebinding of the molecules. The two protein partners are connected to two positions on the J-DNA scaffold and can interact with each other in the absence of significant acoustic force (see Figure 1). Once an acoustic force is applied, the DNA leash is pulled on one end by a bead, rupturing the bond between the two protein partners. The authors measured the changes caused by the rupture of the interacting bond by the extension in the length of the DNA leash when a force is applied.
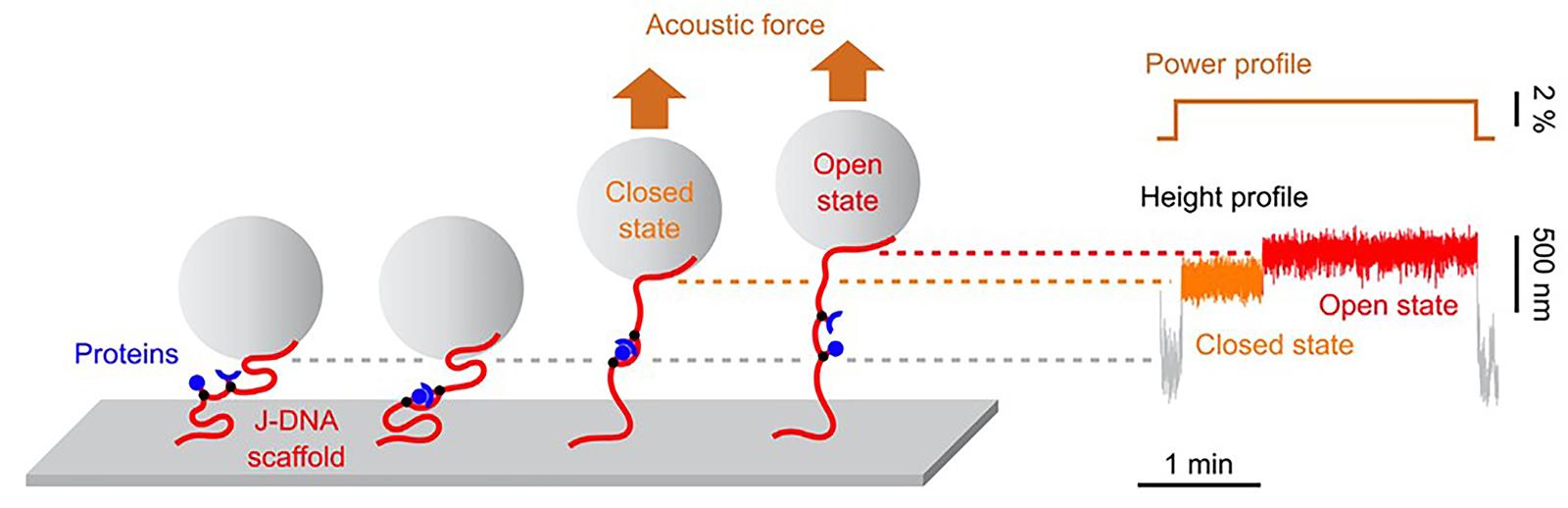
Figure 1. Overview of using DNA scaffolds and acoustic force spectroscopy to measure biomolecular interactions. Left: Beads are tethered to the chamber surface through a J-DNA scaffold (red) maintaining the protein partners (blue) in close proximity via a leash. Bond formation occurs in the absence of significant acoustic force. Upon application of an acoustic force, the difference in extension of the scaffold in the closed and open states allows one to distinguish the unbinding of the protein complex. Right: Corresponding power and height profile of the bead.
One additional aspect of the acoustic force spectroscopy method is high throughput. Traditional single-molecule methods such as optical tweezers, magnetic tweezers, and atomic force microscopy typically look at one interaction at a time, thus requiring a long time to collect reproducible data. By using tailor-made DNA scaffolds as biomolecular leashes, the researchers applied acoustic force on multiple microbeads tethered to DNA leashes. In a single experimental run, they were able to obtain parallel data from multiple such DNA tethers, reducing the typical time and effort needed for single-molecule experiments. With further developments, the researchers hope that this method can be used to study molecular mixtures instead of just a pair of interacting molecules, expanding the application of this method to discriminate molecular mixtures such as polyclonal antibody samples that are frequently screened for clinical and diagnostic uses.
Their study titled “Combining DNA scaffolds and acoustic force spectroscopy to characterize individual protein bonds” was published recently in Biophysical Journal.