
1. Conformational disorder of p21 is linked to function.
Erik W. Martin
Postdoc (Mittag lab), St. Jude Children’s Research Hospital
Over the past several decades, the intrinsically disordered protein (IDP) field has expanded dramatically. The depth of topics covered in the Biophysical Society IDP subgroup symposium over the last several years has highlighted the general acceptance of the crucial roles IDPs play in a wide range of biological functions. This is truly on display in 2021 with a lineup of speakers whose work highlights the greater role of IDPs in the large, macromolecular complexes and assemblies that are essential to life. In light of this, it is often difficult to imagine that it was only within the last decade that it became generally accepted that proteins can exist without stable secondary structure. But naysayers persist. For example, I experienced push-back against this idea fairly recently, in 2017, in a corridor at a recent crystallography meeting. I overheard two scoffing at the notion of an "unfolded protein." This did not feel like the time or place to engage in an argument.
Despite these few hold outs, it is now widely accepted that IDPs are critical to biological function. The goal of this, and articles in this series, is to highlight the seminal work that got us to this point. It might seem like we would have to travel a long way back in time to discover the world before IDPs, but in reality, the first publication to mention the term was only 20 years ago, in 2001. While the notion of a disordered protein had been suggested sporadically in the literature, one of the first concrete examples of a functional intrinsically disordered protein was published in 1996 in PNAS by Richard Kriwacki during his postdoctoral work in the laboratory of Peter Wright at Scripps Research Institute (1). Experimental studies of the mechanisms underlying protein folding was a significant component of Peter Wright’s research program. At the time, functional disordered proteins were primarily just the subject of theoretical discussion. The methods and experience gained studying denatured proteins and characterizing transient secondary structure on pathway to folding provided Peter Wright’s lab with a unique set of tools for studying proteins lacking persistent structure. Confronting these projects was a subgroup in Peter Wright’s lab appropriately labeled the “Large and Difficult Systems” group. Despite moving to San Diego to solve a straightforward structure (and to learn how to surf), this is where Richard Kriwacki found himself.
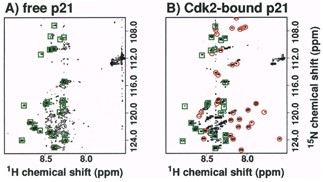
Richard Kriwacki and his coworkers were studying the Cdk inhibitor p21. Their findings can be summarized succinctly. The N-terminal region of p21 effectively inhibits phosphorylation by Cdk at nanomolar concentrations. Despite assumptions that such an inhibitor must be structured, p21 is natively unfolded under physiological conditions. In much the same way that transformative musicians like the Beatles can inspire a generation, the last two decades have seen countless IDP papers that follow this template. To prove that p21 was natively unfolded, Kriwacki and coworkers applied a barrage of techniques traditionally deployed to quantify protein structure, yet they consistently found no evidence of a single, persistent structure. No regions of the protein were protected from trypsin digestion. The circular dichroism spectrum suggested that p21 behaved as a random coil and showed no sensitivity to chemical or thermal denaturation. Finally, the two-dimensional proton-nitrogen NMR spectrum had little chemical shift dispersion in the proton dimension. If a stable secondary structure existed, the local environment of the involved amino acids would have shifted these peaks out of the center of the spectrum.
How then does p21 inhibit phosphorylation? If Cdk2 is added to the p21 NMR sample, the proton chemical shifts are no longer found exclusively within a narrow ~0.8 ppm band. Instead, a specific subset of signals is observed to shift to locations consistent with a folded protein while the remaining signals appear minimally perturbed. The authors had demonstrated that a region of p21 folds upon binding to Cdk2. This paper directly addresses the question that IDP researchers have continued to face in the last 25 years. What is the biological role of folding-on-binding? The authors surmised that an IDP binding partner could engage with multiple targets – a feature that is in line with p21 inhibiting multiple Cdk-cyclin complexes. Furthermore, the authors drew a parallel between p21 / Cdk complexes and Ruth Spolar and Thomas Record's work on DNA binding. They suggested that by requiring multiple favorable enthalpic interactions to counteract the loss of conformational entropy due to folding and binding, IDPs might exchange absolute affinity for specificity (2).
The story does not end here. The notion of folding-on-binding has emerged as a significant theme in IDP research, and the debate continues as to exact mechanistic details. Do these complexes form by induced fit or conformational selection from pre-formed secondary structural elements? "Fly-casting" mechanisms whereby the IDP explores a large conformational space to locate binding partners have been suggested to explain fast binding kinetics, and IDP complexes that contain only minimal folding have been proposed. To highlight how far work on this system has come, I would suggest reading the recent collaborative publication in Nature Communications from the groups of Richard Kriwacki, Claus Seidel, and Peter Tompa on the mechanism of interaction between p27 and Cdk2 / Cyclin A (3).
1. Kriwacki, R. W., L. Hengst, L. Tennant, S. I. Reed, and P. E. Wright. 1996. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc Natl Acad Sci U S A 93(21):11504-11509.
2. Spolar, R. S., and M. T. Record, Jr. 1994. Coupling of local folding to site-specific binding of proteins to DNA. Science 263(5148):777-784.
3. Tsytlonok, M., H. Sanabria, Y. Wang, S. Felekyan, K. Hemmen, A. H. Phillips, M. K. Yun, M. B. Waddell, C. G. Park, S. Vaithiyalingam, L. Iconaru, S. W. White, P. Tompa, C. A. M. Seidel, and R. Kriwacki. 2019. Dynamic anticipation by Cdk2/Cyclin A-bound p27 mediates signal integration in cell cycle regulation. Nat Commun 10(1):1676.